PMxceptional

Working with scientists from the Chicago Zoological Society (CZS) and the Species Conservation Toolkit Initative, we are creating a new version of the current pedigree management software that we are calling PMxceptional. The original PMx software is used in the zoological community to manage breeding programs for endangered animals to retain the maximum amount of genetic diversity over time. PMxceptional will build upon the principles developed in the zoo community to provide genetic and demographic analysis tools appropriate for the characteristics of exceptional plant species.
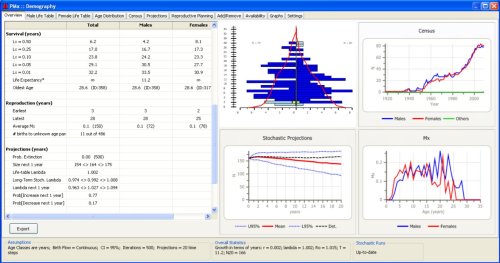
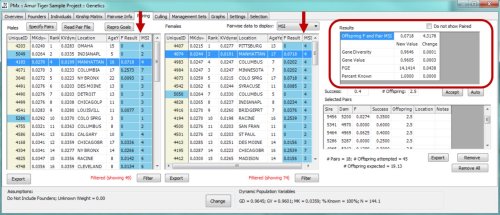
PMxceptional’s analyses will be based on pedigree and demographic data output from the PlantSearch pedigree management module. PMxceptional will be designed to accommodate a wide range of traits not accounted for in PMx, including clonal reproduction, self-fertilization, monoecious breeding systems, sex-change, and the need for demographic models that are stage or size-based rather than age-based. Users will be able to generate information about the ex situ population such as the demographic status (e.g. total number of individuals by sex and reproductive status, population growth parameters, mortality rates) (Figure 2.1) and summary statistics of the genetics of the current population (e.g. genetic diversity, inbreeding), as well as recommendations for which individuals to breed or transfer between institutions (Figure 2.2).
Figure 2.1. Example of an overview from the Demography tab in PMx. Some measures of interest include life expectancy and for males and females, the age structure of collections, and the projection of population sizes over time. From Traylor-Holzer, K. (ed.). 2011. PMx Users Manual, Version 1.0. IUCN SSC Conservation Breeding Specialist Group, Apple Valley, MN, USA.
Figure 2.2. Example of the pairings tab in PMx. In selecting a male and a female (shown with highlighted blue rectangle), the analysis calculates measures of inbreeding and genetic diversity in the offspring, as indicated by red arrows. In addition, the program calculates the overall change in measures of genetic diversity of the population as shown in the results box highlighted with a red square. From Traylor-Holzer, K. (ed.). 2011. PMx Users Manual, Version 1.0. IUCN SSC Conservation Breeding Specialist Group, Apple Valley, MN, USA.
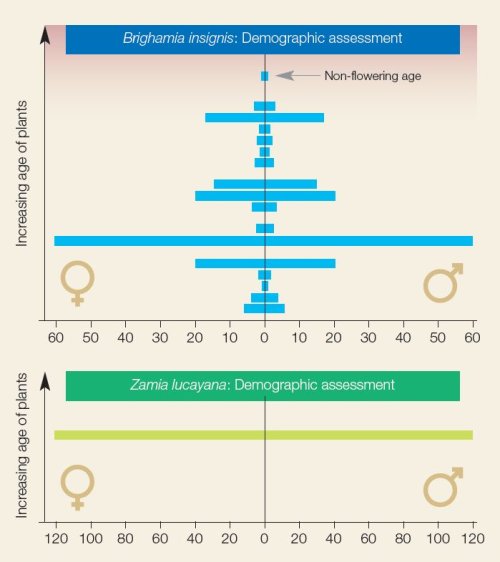
Pilot results Demographic analysis of Brighamia insignis and Zamia lucayana It is important to have individuals of different ages across collections to ensure that there are always individuals of reproducing age available to botanic garden mangers. If all individuals are the same age and become reproductively inactive at the same time, botanic gardens may end up with a species that is no longer able to produce offspring, which could endanger the stability of exceptional species. This is illustrated in a demographic analysis of the Hawaiian endemic Brighamia insignis and the cycad Zamia lucayana (Figure 2.3) The age structure of B. insignis is varied across age classes for males and females, which ensures that younger plants will replace older ones that dye and age. However, all individuals of Zamia lucayana are the same age, jeopardizing the longevity of this collection. This provides vital information to botanic garden managers about the state of a species in their collection and across collections.
Figure 2.3. Age structure from demographic analysis of Brighamia insignis and Zamia lucayana. The number of male (right) and female (left) individuals is shown on the x-axis and the relative age of individuals is shown on the y-axis. From Griffith et al., 2019.
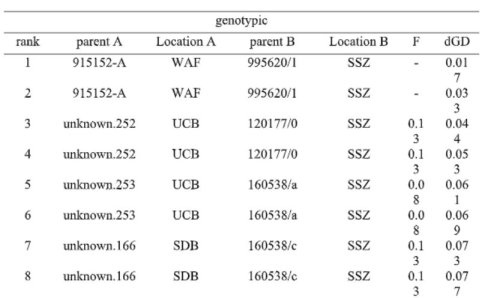
Pairing analysis of Brighamia insignis The most effective way to ensure that botanic gardens retain high levels of genetic diversity for the exceptional species is through breeding individuals that are least related. In using this approach with B. insignis in botanic garden collections across the globe, we identified various breeding pairs at different institutions whose offspring would have the lowest inbreeding coefficient and least positive change in genetic diversity (Table 1). This provides important recommendations about which institutions should be contacted for pollen exchanges when specific individuals are in bloom.
Table 1. First eight ranked breeding pairs of Brighamia insignis from pairing analysis performed in PMx. Rank is based on the lowest mean kinship measured with microsatellite markers. Shown is the rank, the unique identifier of parent A, the location of parent A, the unique identifier of parent B, the location of parent B, the inbreeding coefficient of the pair’s offspring (F), and the change in genetic diversity of population (dGD) as a result of breeding the selected pair. From the master’s thesis of J. Wood, 2019.